Today we will have a blast by travelling ∼270,000 years back in time. And we won’t even need a DeLorean, just our trusty KNIME Analytics Platform. We will return to the Ice Age and examine genetic material retrieved from a cave. We will solve the mystery as to which species the DNA came from.
To investigate this ancient DNA, we will utilize one of the most widely used bioinformatics applications: BLAST (Basic Local Alignment Search Tool). We will do that by running the BLAST RESTful interfaces to submit BLAST searches inside of KNIME Analytics Platform. This post speaks directly to people interested in bioinformatics. At the same time, because it covers the technical aspect of handling asynchronous REST operations, it will also be useful for those in other fields. Thus, if you have an interesting web application you want to use within KNIME - especially one where the resource method execution takes longer - keep reading!
Ok - let's BLAST off!!
Introduction
Bioinformatics without BLAST is like a spaceship without rocket fuel. BLAST is a registered trademark of the National Library of Medicine. The original paper1 has been cited over 70,000 times, not to mention many highly cited follow-up papers 2,3, BLAST is a fast and robust algorithm that finds regions of similarity between biological sequences. The sequences can be DNA or protein sequences from diverse organisms. Biological sequences can be viewed as very large strings like this: AGTCGCAGAGT... The human genome, for example, consists of a sequence comprised of over 3 billion letters. Those sequences change over time, and this drives evolutionary processes (e.g. when one letter is exchanged for another or is removed completely). Hence, if we want to compare many sequences, it is important to use fast and clever algorithms that allow us to find similar sequences even if changes have taken place. BLAST enables a user to compare a query sequence with a huge database of sequences to find the most similar ones.
So what does any of this have to do with our time-travel journey? If we find e.g. a random bone on that journey, we can extract the DNA, and then use BLAST to compare it with a database of DNA sequences and find out which organism the bone belongs to. That’s exactly the sort of problem we will solve now.
Analyzing 270,000 Year Old DNA
In our example, we will analyze DNA extracted from an archaic femur (thigh bone) from the Hohlenstein–Stadel cave in southwestern Germany, which is at least 270,000 years old. We use part (~50%) of the DNA sequence published last year by a research team led by scientists from the Max Planck Institute for the Science of Human History and the University of Tübingen4. More specifically, we will investigate mitochondrial DNA (mtDNA). Sequences of mtDNA sequences are relatively short and, since they evolve faster than nuclear genetic markers, they accumulate differences comparatively quickly. This makes the analysis of mtDNA one of the most important tools for phylogenetics and evolutionary biology.
We will use KNIME Analytics Platform to compare that mtDNA sequence with a huge set of DNA sequences from diverse organisms. For that, we create a workflow that receives a sequence as input, queries BLAST and shows the BLAST results in an interactive table (see Fig. 1).

Figure 1: BLAST workflow. The workflow receives an input sequence, queries BLAST, and subsequently shows the BLAST results in an interactive table. The views are accessible via KNIME WebPortal or via the node view.
We compare our sequence to the extensive Nucleotide database, a collection of sequences from several sources, including GenBank, RefSeq, and PDB. In July 2018 RefSeq alone covered genetic data from more than 81,000 organisms. Comparing the input sequence with the vast number of sequences in the database can take a while, hence the BLAST RESTful interface is handling queries asynchronously.
Handling Asynchronous REST Operations
Calling a web application synchronously implies that you have to wait for the response to return before you can continue. In cases where the response takes longer this blocks your workflow. In contrast, asynchronous operations are non-blocking, i.e. client execution continues immediately after the request, and the response is then processed later, by a different execution context or thread5.
Handling asynchronous REST operations introduces some challenges, as we receive a return value even when the execution is not finished. What that means for us here is that after starting our BLAST job our workflow will repeatedly call the web service to check whether or not it has finished. This workflow is contained in the metanode “Query BLAST” and is depicted in Figure 2. We first create a new resource using the PUT method. We then extract the “Request Identifier” (RID) and the “Request Time of Execution” (RTOE). Afterwards, we build a loop where we keep waiting RTOE seconds until the status of the request equals READY. Now that we know that the BLAST results are ready for retrieval, we can fetch them via a final GET request.
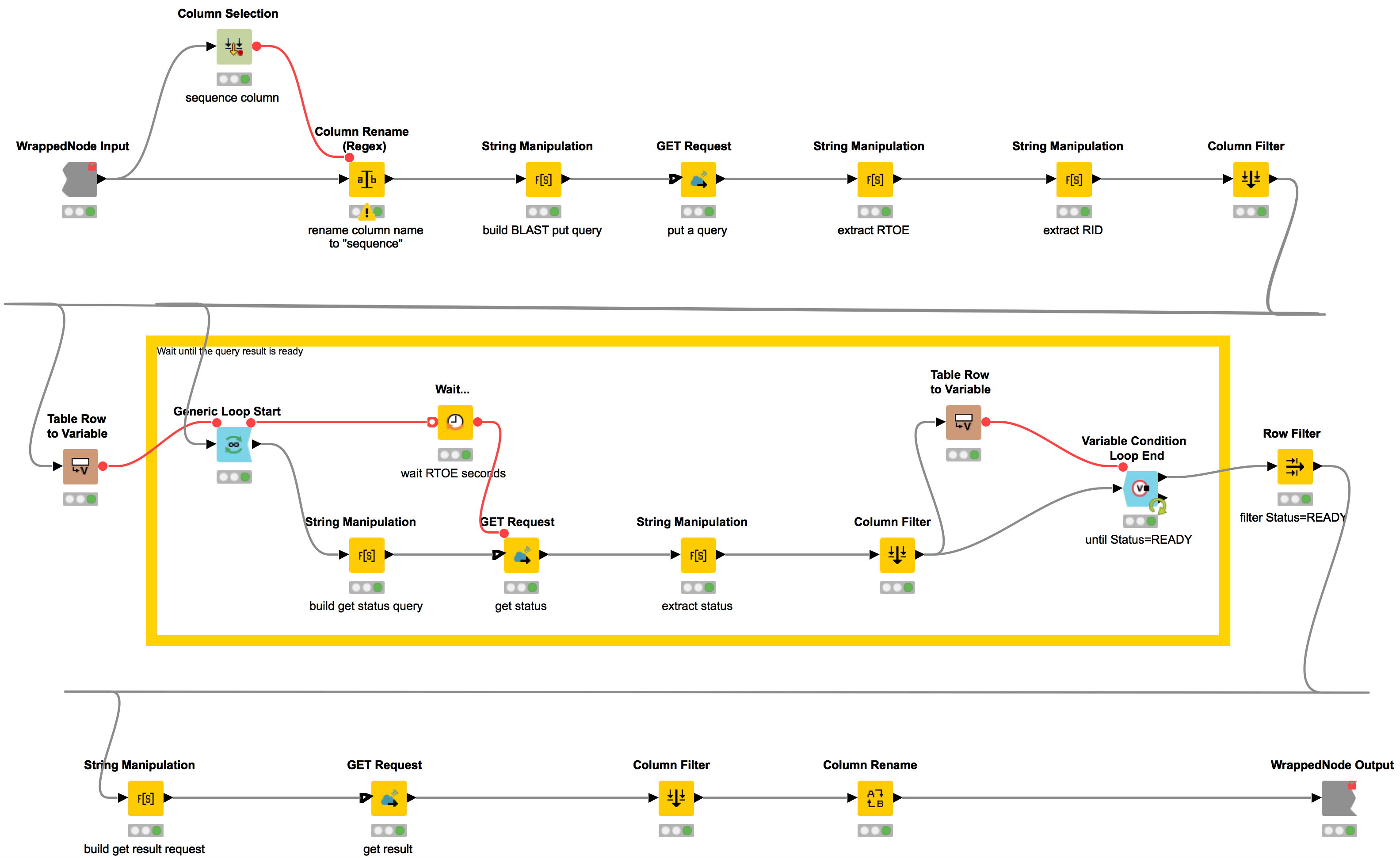
Figure 2: Retrieval of results from an asynchronous GET Request: First, we extract the 'Request Identifier' (RID) and the “Request Time of Execution” (RTOE). We then build a loop where we keep waiting RTOE seconds until the status of the request equals READY and subsequently retrieve the BLAST results. This workflow is contained in the metanode “Query BLAST”.
BLAST Result
The BLAST query gives us an XML document, which we parse using the XPath nodes in KNIME. We extract the description of the hit, the accession ID, the length of the sequence, the score, and the e-value. The score adds up the positions where the two sequences are the same and penalizes those where they differ. The e-value describes the expected number of hits with an equal or higher score one would expect to see by chance when searching a database of a particular size. The lower the e-value (the closer it is to zero), the more “significant” the match is. In addition, we collect the field identity, which signifies the extent to which two sequences have the same residues (letters) at the same positions. By dividing by the length of the sequence and then multiplying by one hundred we receive the percentage of sequence identity that is shared between the input and the hit sequence. For details of the calculation and interpretation of the extracted results, see the BLAST Glossary.
Now it's time for the drum roll… What will we find? Who did the mysterious bone belong to?

Figure 3: JavaScript Table View of the BLAST Result. The Description, score, e-value, accession, and Identity_percent are displayed. The e-value describes the expected number of hits with an equal or higher score and Identity_percent signifies the percentage of sequence identity that is shared between the input and the hit sequence. This is the output of the metanode “Show BLAST results”.
Our top hit, with 100% identity and a significant e-value, is “homo sapiens neanderthalensis mitochondrion”. This is the DNA that was submitted by the scientist from the Max Planck Institute. The following hits also manifest Neanderthal mtDNA from different archaeological sites. Our bone, conclusively, was a Neanderthal bone! Different isolates of homo sapiens mtDNA are also among the significant hits with an identity of ~98.9% showing once more that we are not as different from the Neanderthals as we might think.
Summary
We have showed how to blast off with the KNIME rocket ship, traveled in time back to the Ice Age and investigated ancient DNA. We revealed that the DNA we found was Neanderthal mtDNA and also compared the Neanderthal sequence to sequences from modern humans.
To accomplish all of that, we used an interactive KNIME workflow that is able to handle asynchronous REST operations. This workflow makes use of the RESTful interfaces offered by the NCBI to programmatically submit BLAST searches. You can use this workflow as a template to query all kinds of RESTful interfaces. In addition, the workflow can be easily deployed to the KNIME WebPortal, so the end user does not even have to know about the rocket science under the hood.
References
1. "Basic local alignment search tool. - NCBI - NIH." https://www.ncbi.nlm.nih.gov/pubmed/2231712. Accessed 20 Jul. 2018.
2. "Gapped BLAST and PSI-BLAST: a new generation of protein database ...." https://www.ncbi.nlm.nih.gov/pubmed/9254694. Accessed 20 Jul. 2018.
3. "BLAT--the BLAST-like alignment tool. - NCBI." https://www.ncbi.nlm.nih.gov/pubmed/11932250. Accessed 20 Jul. 2018.
4. "Deeply divergent archaic mitochondrial genome provides ... - Nature." 4 Jul. 2017, https://www.nature.com/articles/ncomms16046. Accessed 20 Jul. 2018.
5. "Managing asynchronous Web services interactions - IEEE Conference ...." https://ieeexplore.ieee.org/document/1314726/. Accessed 15 Aug. 2018.
6. "BLAST Glossary - BLAST® Help - NCBI ...." 14 Jul. 2011, https://www.ncbi.nlm.nih.gov/books/NBK62051/. Accessed 23 Jul. 2018.
