If the diagnosis of diseases affecting agricultural crops is delayed or inaccurate, it can cause strong environmental impacts and significant economic losses. The over- or under-use of chemicals due to misdiagnosis of crop diseases can lead to resistant pathogen strains, increased production costs, increased environmental and health impacts, or even a potential outbreak and diminished harvest.
Currently, plant disease diagnosis is based on human scouting, which is very time-consuming and expensive. Automated imaging processing and the application of machine learning has the potential to significantly speed up plant disease diagnosis.
In this blog post, we create a single machine learning model to predict two apple foliar diseases, rust and scab. Both are caused by fungi and lead to reduced fruit quality and premature defoliation. Rust can be identified by yellow leaf spots which eventually turn a bright orange-red color. Scab forms pale yellow or olive-green spots on the upper side of the leaves. The provided data contains real-life symptom images of the two diseases, with variable illumination, angles, surfaces, and noise, as well as images of healthy leaves1.
We are taking advantage of the pretrained Resnet50 model2, which is a residual deep learning neural network model trained with 50 layers in Python. Its original purpose was to classify 1,000 different objects, and it has been trained on millions of images to do so.
We will load, adjust, and train the model in KNIME Analytics Platform to make it applicable for our problem, which is to identify rust and scab. The workflow will demonstrate how to load and pre-process the images to be suitable input for Resnet50, how to train selected layers, and how to evaluate the model’s performance.
You can download our Plant Disease Prediction workflow from the KNIME Hub to try out yourself. Please be aware that it is very large. You can find the input images in the Plant Pathology Challenge 2020 dataset.
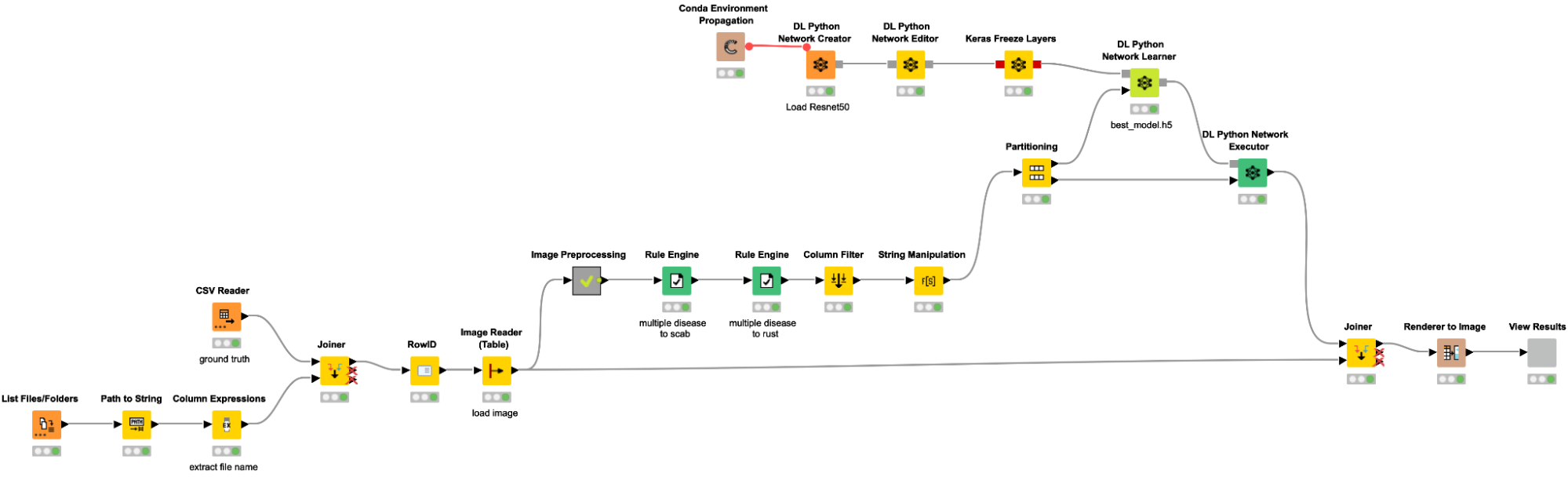
Loading images and data into KNIME
As a first step, we are loading our data into KNIME. The workflow snippet below creates a table with the path to all images, adds the label based on the file name along with an additional csv file, and reads the images with the Image Reader node. We are importing each image file and joining them with the training labels for each image, which are stored in a separate train.csv file (possible labels include “healthy,” “scab,” “rust,” and “multiple diseases”).
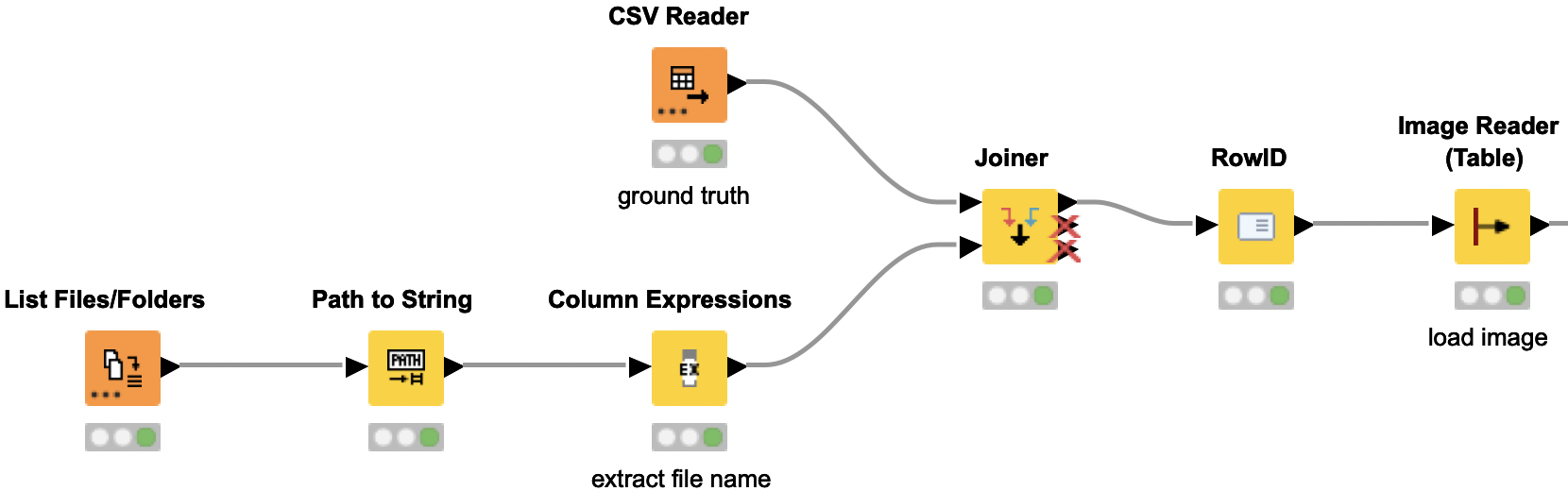
Image preprocessing to provide the suitable format for Resnet50 model
Now we need to preprocess the images to provide them in the required input format for the Resnet50 model. The initial dataset contains duplicate images with different labels — i.e. an image that is labeled once as rust and once as scab. We remove those duplicate images with different labels, as this would interfere with the models training. Next we are resizing the images and splitting them by the RGB channels. Each channel is then normalized in the same way as the images for the original Resnet50 model by subtracting the mean values for each channel. Then the channels are merged back into one image.
Redefining the problem: Are rust or scab present?
Our original task was to identify the four classes: “healthy,” “scab,” “rust,” and “multiple diseases.” However, the classes are very unbalanced. There are only 91 images of leaves with both diseases, whereas there are around 600 each for scab and rust. Therefore, we are redefining the problem into two binary classifications. We want to simply identify whether rust is present or not, and whether scab is present or not. Based on those two predictions, we can easily conclude if a plant is healthy or has multiple diseases (rust and scab).
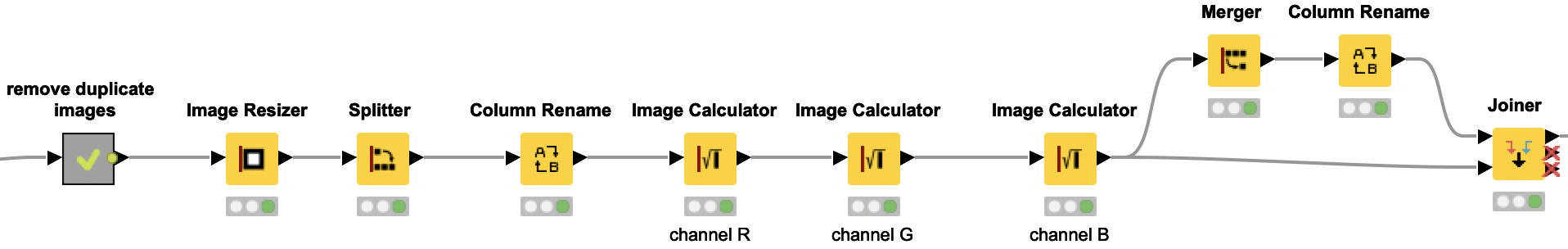
Training and applying the Resnet50 model
The pretrained Resnet50 model is loaded from Keras using the DL Python Network Creator node. However, we only retrain the last two thirds of the network. We will keep the weights of the first third of the layers by freezing them. In the DL Python Network Editor node, we add two more dense and one dropout layer to the original model to get the correct output format, which is now two output values after we redefined the problem (i.e. solving two problems, predicting the presence of scab and rust). With the Keras Freeze Layers node, we are freezing the first third of the layers and only retrain the last two thirds of the layers, as described above.
DL Python Network Learner Node
We are using the DL Python Network Learner node. This way, we can simply use the Python code below to train our model. First we need to load and prepare our training data. To do so, we are converting the KNIME image data type to a numpy array. We are combining the rust and scab column to one combined y-train. Since our data is very imbalanced, we are also calculating the class weights to later provide them for the model training.
Next we are splitting our training data into a training and validation set. The performance of the model on the validation set is used to determine the early stopping. This way, our model fitting is automatically stopped if the performance on the validation set is not increasing after five iterations, — or even worse, decreasing. We are using the keras early stopping function and the checkpoint callback function to define the criteria for an early stopping. While we are training the model, we are writing the model to a file and “remember” the validation accuracy. If the validation accuracy is increasing in comparison to the previous iteration, we are overwriting the model with the newly trained model. If the validation accuracy does not increase for the last 5 batches, the model training is stopped. Additionally, we are using the Keras Image Data Generator for data augmentation. This way we are modifying existing images and creating new ones by simple changes to the image like rotating, flipping or shifting. This adds a little bit of noise to our data and avoids overfitting.
Similarly to the DL Python Network Learner node, we are using the DL Python Network Executor node to use Python code to apply our model on the test data. We are loading the test images and the model that we trained in the previous step to get the predictions of whether the leaf in each image shows signs of infection with rust and/or scab. The predictions are then added to the input table.
Evaluating the model performance interactively
In a final step, we use an interactive view to evaluate the performance of our model. We simply use two confusion matrices to evaluate the performance of the model for each of the two diseases. The interactivity of the color-coded table view in combination with the confusion matrices makes it easy to explore the prediction of the model and to quickly grasp and interpret the model predictions.
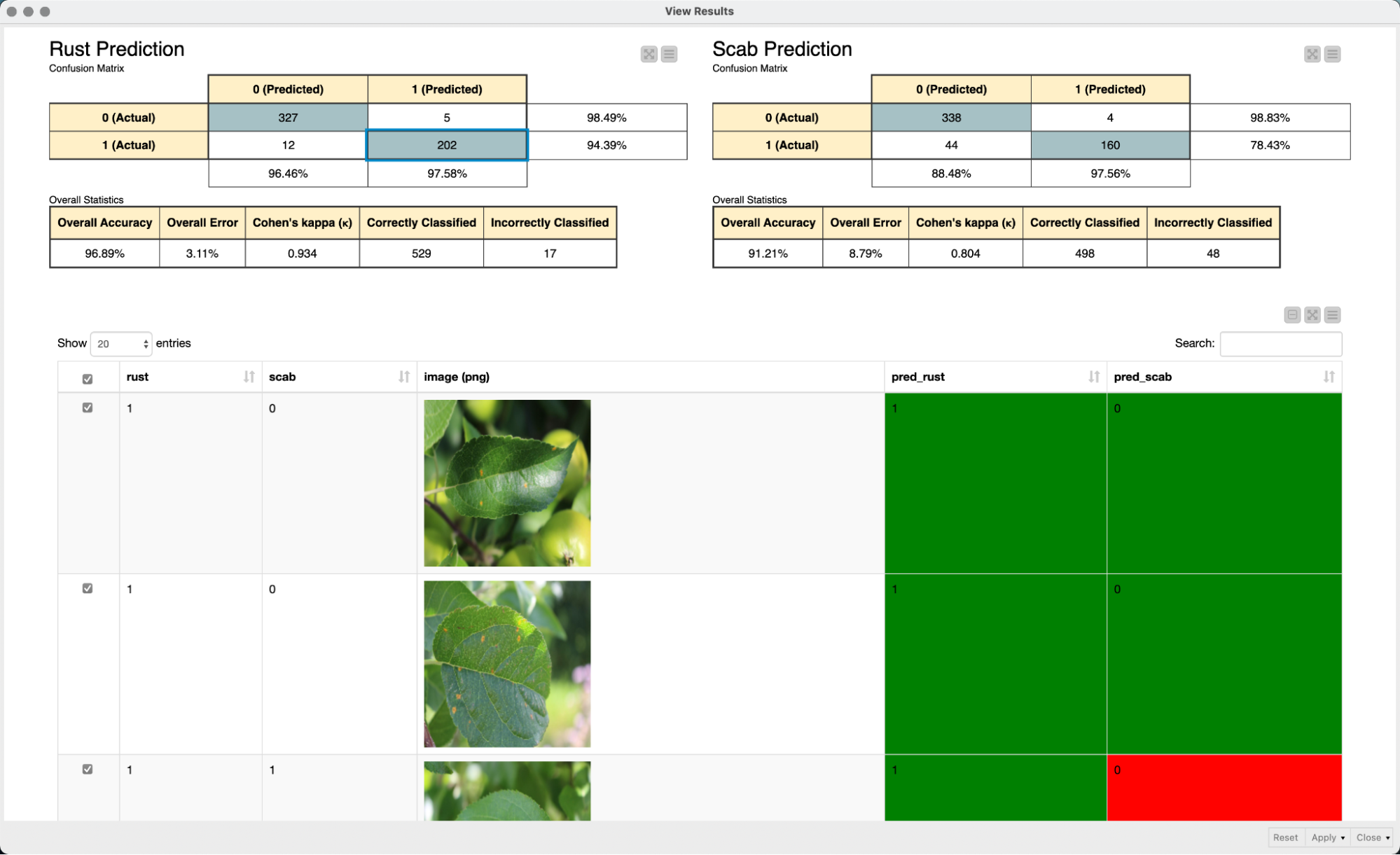
Summary of the steps for plant disease diagnosis
In this post, we have shown how to load and modify a pre-trained Keras model in KNIME Analytics Platform to classify images of crop diseases. The steps included image pre-processing, adjusting the pre-trained model, and only retraining part of the layers to adapt to our problem. To improve the performance of the workflow, we used the new Columnar Table Backend.
