Many life science companies have important digitalization initiatives that incorporate end-to-end integration. The goal of such initiatives is to improve experimental reproducibility, reduce errors, increase productivity, and enhance the generated data through contextualization. However, the effort required to implement these is high, due to contending with inconsistent infrastructures as well as a diverse set of integration interfaces. This hinders an effective implementation; it often seems easier to stick to doing everything manually. A solution to this problem is to integrate standards and to simplify end-to-end integration.
Standardized lab automation with KNIME Analytics Platform
In this blog post, we show how we integrated the SiLA standard (Standardization in Lab Automation) in KNIME Analytics Platform through a collaborative effort of siobra, Biosero, HDC, KNIME, and the SiLA consortium. SiLA enables the automation and digitizing of scientific laboratories through free and open data standards and systems communication.
We utilize the SiLA integration in two use cases that automatically analyze
These workflows are useful beyond these two use cases. The SiLA standard can be used in a lot of different scenarios, and the workflows easily adjusted and applied to diverse projects. With the demonstrated integration we enable lab scientists to perform their own analyses by building reproducible workflows to automatically retrieve their data, blend them with other data sources, and carry out different kinds of post-processing.
Video demonstrating automatic retrieval and analysis of lab data
Check out this video to see the workflows we built for the two uses cases in action. The video demonstrates how KNIME workflows can automatically retrieve and analyze laboratory data in one simple and intuitive environment for end-to-end data science.
SiLA for open connectivity in lab automation
The SiLA Consortium is a non-profit membership organization that brings together lab users, vendors, and start-ups to define standards that permit information interchange in the lab. SiLA’s mission is to establish international standards that create open connectivity in lab automation. It provides a framework for free and open system communication and data standards to connect scientists with the data that matters, and it is supported by some of the biggest and most innovative names in the industry. It is built using a message-driven architecture based on HTTP/2, thereby supporting the delivery and management of data from a wide range of laboratory devices. Devices expose their properties and commands (i.e., operations) to the network via the so-called SiLA 2 server. A SiLA 2 client can then establish a network connection to the server and initiate command execution on the device 1.
SiLA and Biosero were at #SLAS2021 Digital International Conference and Exhibition sharing more about the collaboration and demonstrating 'the power of standards in action'!
SiLA and Biosero were at #SLAS2021 Digital International Conference and Exhibition sharing more about the collaboration and demonstrating 'the power of standards in action'!
sharing more about the collaboration and demonstrating 'the power of standards in action'!
How does SiLA integrate with KNIME?
The SiLA consortium and its members maintain reference implementations of the SiLA 2 standard for various programming languages. Among others, an implementation for Java is available at https://gitlab.com/SiLA2/sila_java under MIT license. Since the sources are publicly available, siobra was able to repackage this reference implementation and implement a light-weight wrapper that can readily be used from KNIME Analytics Platform. Early in the project we decided to go for a proof-of-concept implementation that uses Java Snippet nodes utilizing the aforementioned wrapper. To make this usable (but also adaptable) by others, we have encapsulated this functionality into shared components. Please note that in order to minimize the size of the components, the JAR file containing the wrapper is available separately. This has also allowed us to iterate rapidly on the integration together with the other collaborators, e.g. by replacing one instance of the wrapper that is used in all components. It furthermore allows tech-savvy users to use components as blueprints for connecting to their lab devices via SiLA (without the need to recompile and package sources).
We have created three shared components to handle different tasks: one to extract binary data such as images from the SiLA Server, one to retrieve XML data, and one to send the results back to the SiLA Server. The three components can be found on the KNIME Hub for easy reuse. Click the links to access and download the components from the KNIME Hub.
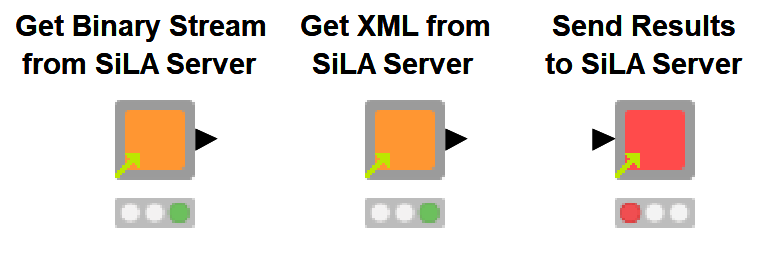
Fig. 1. Shared components to retrieve binary (e.g. images) and XML data from SiLA Server and send the results back to the server.
Use Case: Automatic Analysis of Microplate Data
The first example workflow demonstrates how you can use the new shared components to retrieve plate readouts (e.g., fluorescence) as XML from a SiLA server, process the data with KNIME nodes, and send computation results back to the SiLA server. You can download the workflow “Get Data from SiLA 2 Server and Send Back Results” from the KNIME Hub here.
The workflow comprises four simple steps as can be seen in figure 2.
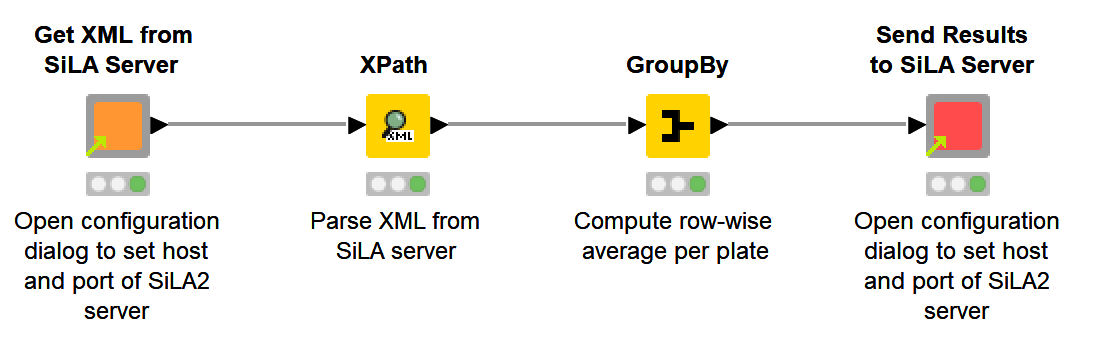
Fig. 2. Workflow to prompt measuring data on a microplate, extracting and processing the data and sending the results back to the SiLA Server.
1. Get XML from SiLA Server
We use the shared component to retrieve microplate readouts. A connection to the SiLA 2 Server is established based on the given host name and port and performing the measurement is triggered. The data is then sent back to KNIME as XML.
2. Parse XML
The retrieved data is parsed into a table using the XPath node, which allows to perform XPath queries to create a data table in an intuitive way.
3. Process the data
In this simple example, we perform a row-wise average per plate using the GroupBy node to process the data. More complex operations including interactive visualizations would be possible in this step as well.
4. Send Results to SiLA Server
In the last step, we send the data back to the SiLA 2 Server using the provided shared component. In addition to the host name and port we also have to configure the column with keys and values.
Use Case: Automatic Analysis of Imaging Data
The second example workflow demonstrates how to analyze data from an imaging experiment that comprises multiple, potentially multi-channel or time-resolved, images. That is, instead of well-structured XML from a plate reader, we receive binary data in the form of images from a SiLA Server. In this example, we do not get each image as an individual file, but rather collected in a compressed archive (e.g., ZIP). Once the contents have been extracted, we use the KNIME Image Processing Extension to derive information from the images.
You can download the workflow “Get Images from SiLA 2 Server and Send Back Results” from the KNIME Hub here.
In figure 3 below, you can see the entire workflow, comprised of the following steps:
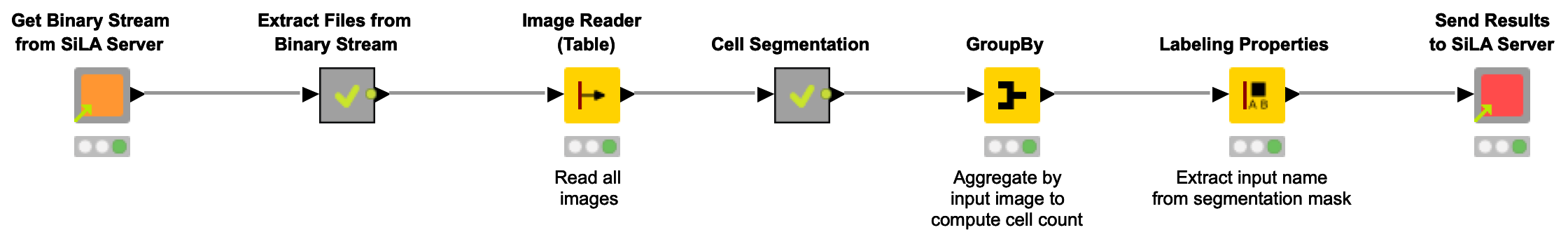
Fig. 3. Workflow to receiving images, extracting cell counts, and sending the results back to the SiLA Server.
The steps of the workflow are as follows:
1. Get Binary Stream from SiLA Server
Request and receive a binary stream from SiLA Server. This automatically creates a Binary Object column that can be handled with generic KNIME functionality (see below).
2. Write Binary Stream to Archive File and Extract
Take the Binary Object column and write the content into a file in a dedicated temporary directory. Since we expect the binary stream to contain an entire ZIP archive, we create and extract such an archive. A table with file paths (in this case: images) is returned from the metanode.
3. Read Images
Use KNIME Image Processing to read all received images into a column of a dedicated Img type.
4. Cell Segmentation
This metanode implements a simple, intensity-based cell segmentation approach. That is, the cells in each input image are identified based on the fact that they show an increased intensity in comparison to the background of the images.
5. Computing Cell Count per Image
Here, we aggregate information from all input images in order to compute the number of cells per image. This information can, for instance, be used to determine if the cells have reached the desired density to continue with the experiment
6. Send Results to SiLA Server
Using the aforementioned component, we send back the cell counts per image.
Summary: Reproducible Workflows for Automatic Retrieval and Analysis of Lab Data
We learned today how to create reproducible workflows that automatically retrieve and analyze laboratory data. All this can be done in one simple and intuitive environment for end-to-end data science.
The collaboration of siobra, Biosero, HDC, KNIME, and the SiLA consortium worked together to integrate the SiLA standard in KNIME Analytics Platform. In this article, we showcased the new SiLA integration in two use cases: one automatically analyzes microplate data and the other automatically analyzes imaging data. You can now simply download the shared components and workflows and apply them to your own data. Try them out yourself!
References
1. Porr, Marc, et al. "Implementing a digital infrastructure for the lab using a central laboratory server and the SiLA 2 communication standard." Engineering in Life Sciences (2020).
