Dr. Anthe Janssen is an assistant professor (tenured) in the field of computational chemical biology at Leiden University, The Netherlands. He uses KNIME in an MSc-level course that teaches the basics of cheminformatics and machine learning as well as in supporting research projects from a computational perspective. In this article, we hear why he chose KNIME as his teaching tool.
A few years ago, I joined the faculty of the Leiden Institute of Chemistry. Specifically, the division that focuses on chemical biology to study biology using chemistry tools and techniques. The division exists somewhere on the interface of organic chemistry, life sciences, biology, and pharmacology.
The division is quite experimentally focused, which is reflected in its master’s programs.
I was asked to set up a new course introducing master’s students to computational concepts useful in chemical biology. This presented me with a sizable challenge: How do I familiarize experimentalists with cheminformatics and machine learning?
My teaching challenge and why I chose KNIME
Most of our current master’s students have not had any meaningful programming or data science courses, and apart from a foundation in theoretical chemistry focusing on quantum chemistry, they have no computational background. The course had to be designed to appeal to most students but more importantly be doable for them.
It also had to introduce concepts such as protein modeling, deep neural networks, molecular dynamics, and free energy perturbations. We also wanted “practicals” incorporated into the course to show students that they can easily apply the concepts , encouraging them to consider the techniques in their future research.
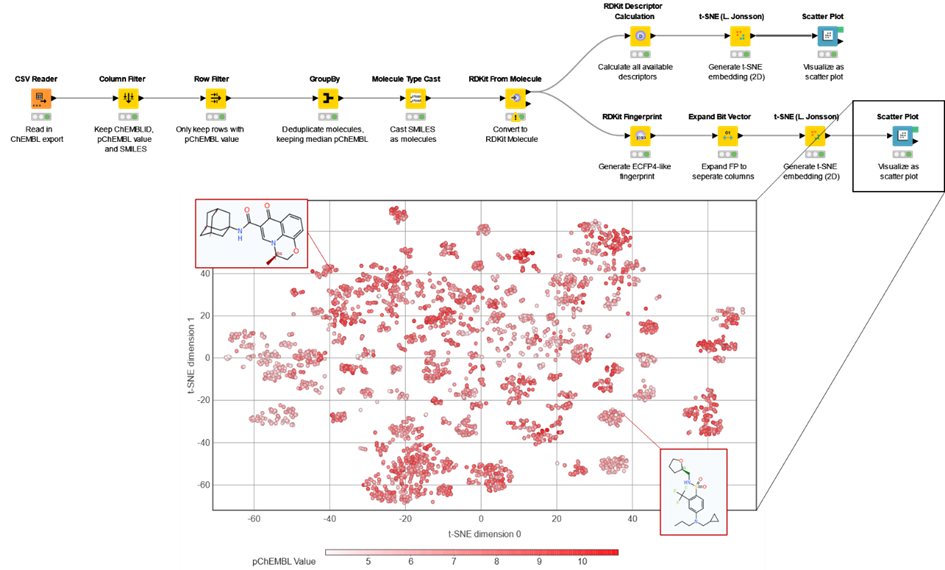
In the final design of the course (Figure 1), we decided one of the three practical sessions would be taught entirely in KNIME.It would apply concepts from cheminformatics and illustrate the power of machine learning, all within the user-friendly workflow views. I will detail why we made this choice below.

In the lectures leading up to the KNIME practical, we cover a big chunk of cheminformatics and a basis in machine learning. We want the students to have an understanding of chemical diversity and how to assess that using molecular fingerprints.
We also teach them what random forests and deep neural networks are, how they work, and how to assess the performance of a model. These topics we also apply in the practical. In lecture 6, we also spend some time introducing the basics concepts of KNIME to prepare for the practical.
Quickly and easily build a workflow doing "actual stuff"
The first part of the task involves using KNIME’s RDKit integration. The students build a workflow from scratch to experience how this process works. They are given detailed instructions on which nodes they’ll need. They learn how to load a set of molecules from ChEMBL (also discussed in the lectures), parsing them as the correct datatype into RDKit, and using that to calculate ECFP-like fingerprints. These are then used to generate a scatter plot of the “chemical space” from a t-SNE embedding.
The two main takeaways of this part of the practical are:
-
Illustrate the use of abstract techniques introduced in the lecture
-
Show them how to get a workflow up and running without coding
TeachOpenCADD: Immediate hands-on machine learning
In the second part of the practical, we want the students to get hands-on experience with training machine learning models. For the machine learning aspect, we use TeachOpenCADD-KNIME. TeachOpenCADD is a teaching platform that includes a set of workflows, published originally in 2019. It is still actively maintained and expanded upon. The licensing terms allow us to adapt the freely provided workflows to our needs.
Initially, we removed everything but sub-workflows 1, 2, and 7: ChEMBL data acquisition, molecular filtering, and ligand-based machine learning. This meant that students still had to drill down into metanodes and components to find the settings they needed to adjust, such as the number of layers of the DNN and the number of trees in the Random Forest models. In subsequent years, we’ve streamlined all these settings through component configuration options. Now, it is much simpler to get started and train neural networks to predict bioactivities.
Readily available learning materials
During the lectures, we don’t have the time to discuss details about the software the students will be using in the practical assignments. We want to focus on the concepts and workings of the data science techniques, their power, and their shortcomings.
Therefore, in the lecture before students begin the practical, I explain only the very basics of KNIME: the concept of workflow-based analysis tools as well as a few slides about what KNIME nodes are and how to execute them. The intuitive interface is easy to understand and I can point students to a vast range of online instructional videos about KNIME for them to dive deeper.
This allows me to focus our lectures on the content, while still giving the students a good way of getting familiar with KNIME.
Note. A wide variety of teaching materials is readily available through the KNIME Educators Alliance, which teachers can freely join. The videos used in our course directly come from the teaching materials repository, accessible through the Educators Alliance.
Will I stick to using KNIME to teach data science?
As mentioned, our future students will be increasingly adept at working in a code environment. Not only are our bachelor’s programs introducing courses around this topic, school systems are also exposing students to programming languages from an early age. Everything we do in our KNIME-based practical, such as TeachOpenCADD, can be done in Python as well. However I see good reason to stick with our current course design.
Our course is about getting students familiar with the concepts and technologies. Through the low-code environment, they can focus immediately on the analytics, rather than learning the coding language. With this hands-on experience we maximize their chance of understanding what they are doing quickly and seeing the added benefit.
Besides, KNIME is incredibly flexible software. If students are looking for a way to solve a specific task, a phrase they’ll often here is: “You know there is a node for that!’ And if there isn’t a KNIME-native node to perform the task, the KNIME-Python integration enables you to incorporate some Python into your workflow and add the functionality yourself.
What’s not to stick to?
