An ever increasing body of scientific literature is documenting significant advances in healthcare research and novel treatments. However, these valuable insights often get lost in the exponentially increasing volume of published studies.
A single query often results in a huge number of publications, all of which needs to be carefully read through. This quickly becomes an overwhelming task. Narrowing the query parameters to bring down the resultant information to a manageable number runs the risk of the query being too specific and leading to a few or even no results. These narrow queries can also be a significant barrier to incidental findings that may be of relevance to the question under investigation.
A simple approach allowing the querying of two overlapping but different sources such as PubMed or Semantic Scholar in parallel, and using highlighting to quickly identify the relevant information will not only save time, but go a long way towards minimizing information overload while keeping a query broad enough to allow incidental findings.
In today’s blog we will demonstrate how such an outcome may be achieved using the open source KNIME Analytics Platform.
We created two example workflows to automatically extract abstracts from publications.
-
Example #1 investigates Adverse Drug Events (ADE’s) in the context of the ethnic background of a patient: Download workflow: Search for Ethnicity Related Adverse Events of a Drug.
-
Example #2 investigates relationships of particular genes and diseases. Download workflow: Tagging Genes in Disease Related Publications
With both workflows we use ontologies, dictionaries, or web services to get a list of terms or synonyms to help with the search. We also tag terms of relevance in abstracts and highlight them in a manner to make the relevant information much easier to find. Let's have a look at the two workflows.
Example #1: Investigating Adverse Drug Events & Ethnicity
Genetic polymorphisms in enzymes involved in ADME (absorbance, distribution, metabolism and excretion) have been demonstrated to affect drug response as well as ADE’s. Specific polymorphisms in these enzymes have been demonstrated to be differentially concentrated among different ethnic groups. This natural phenomenon leads to ethnicity dependent variations in drug responses and ADEs.
Often, the balanced representation of different ethnic groups in clinical trials are difficult to achieve. Thus medications once approved are used to treat ethnic groups on which the medication are not tested. As such, a drug that is safe for the majority may have a differential response or ADE in a different ethnic group due to inherent variations in polymorphisms among ADME genes.
Early identification of such rare adverse events will likely be detailed in clinical reports with one or a small number of patients. Thus such information is often lost in the vastness of published literature unless a routine ADE surveillance effort is implemented that specifically investigate ADE’s in the context of ethnic backgrounds.
In the workflow we have created, we look into the example of Carbamazepine and the Stevens-Johnson Syndrome which is a serious skin disorder. Let’s check if there is literature available that would suggest a dependency on any kind of ethnic background of patients.
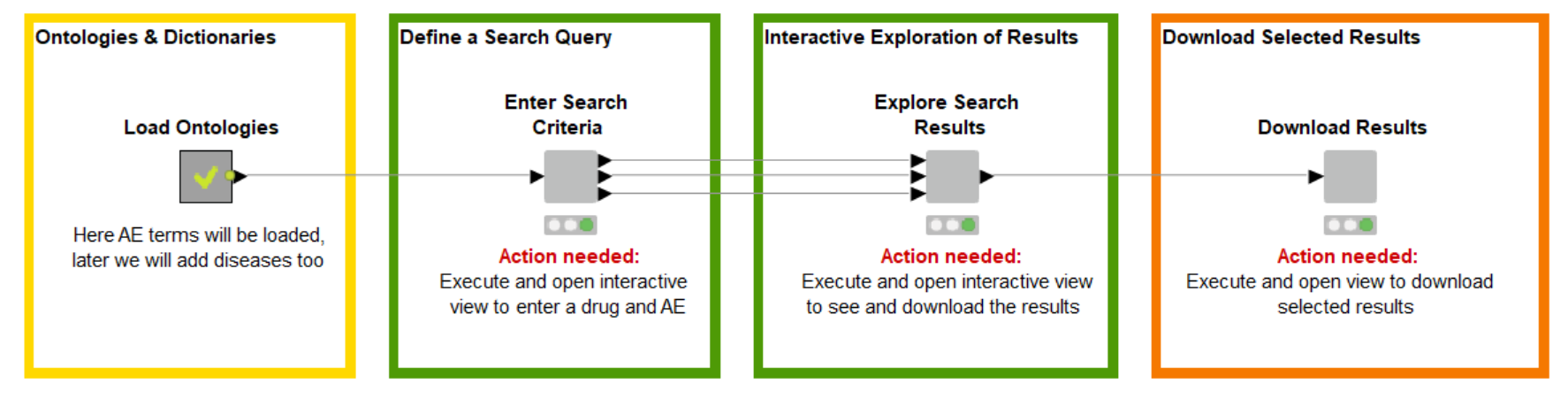
Create Query for Web Services
With this outline in mind we created a workflow that allows the user to search within two different literature sources for publications that relate to the search query using only one tool – the open source KNIME Analytics Platform.
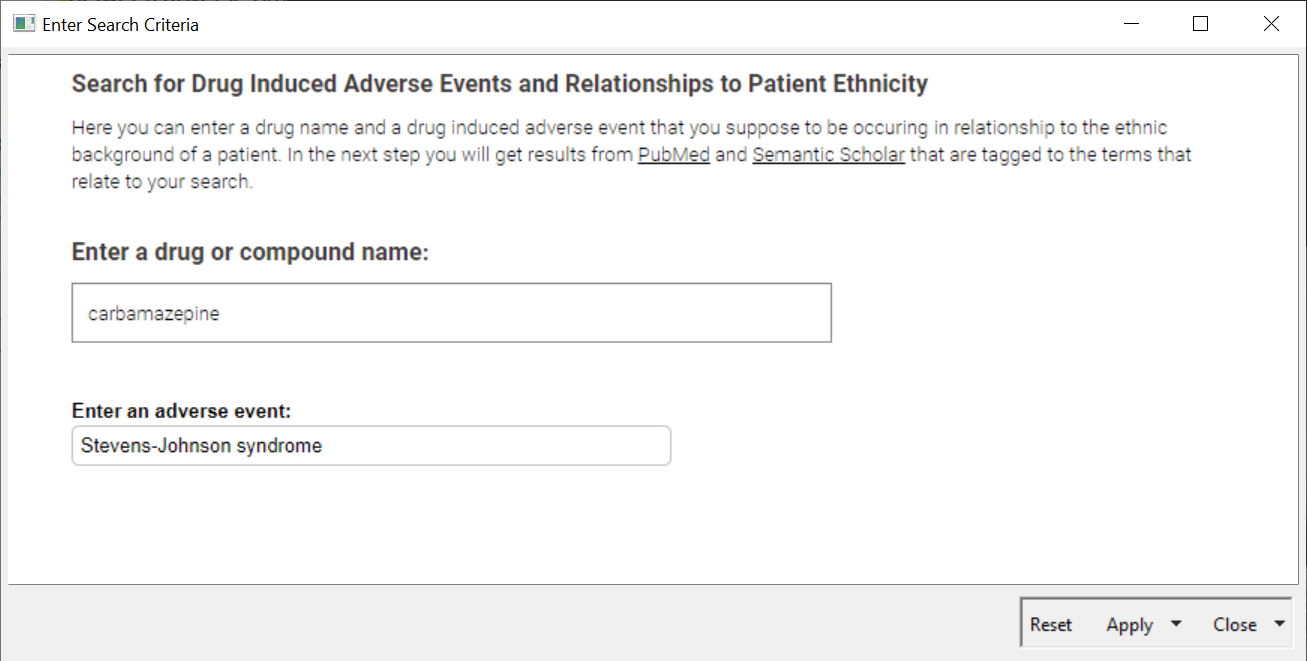
In the first step the user can add a name of a drug and an adverse event into an interactive view of a component (See Fig.2). It's not only two simple text fields – we added more functionality in the backend that to improve the serch. The adverse event field is made of an Autocomplete Widget node and, based on the Ontology of Adverse Events, proposes terms directly when the user is typing first characters. Nevertheless, if a term isn't available, it will use what is entered to create the query.
Depending on the author of a publication, different names of a particular drug that is described is used. Sometimes the compound name or the marketed drug name could be entered. For example: some people would say Aspirin, others Acetylsalicylic Acid. To make sure we catch as many publications as possible, we collect synonyms for the entered drug using the ChEMBL Web Service.
Finally, in order to improve the search, we added two different sources to look for publications. The first is PubMed that can be called using the Document Grabber.
In our research to see if other APIs are available, we stumbled upon Semantic Scholar which is an invention of the Allan Institute and contains a really easy to use web service. Please read through the license and the API documentation, before you start using it in the workflow.
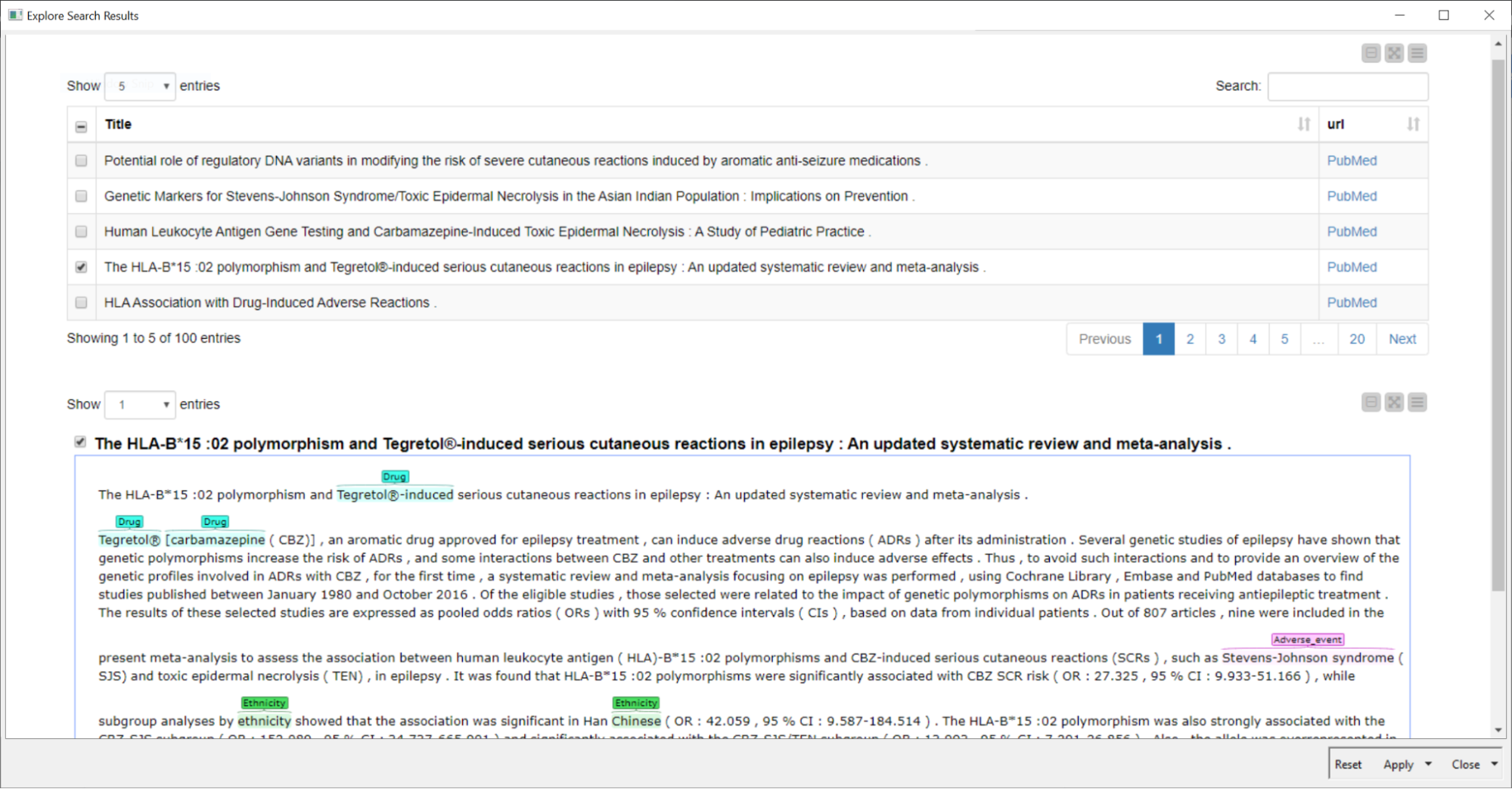
Remember the time when publications were printed and you used to sit with a text highlighter or marker to mark the important parts? This is done here automatically on the abstracts using the Dictionary Tagger and Tagged Document Viewer nodes.
Check the Results
So let’s have a look at our example Carbamazepine & Stevens-Johnson Syndrome to see if there is literature available that shows a relationship between both terms and the ethnic background of patients. Indeed there is a lot of information already available (See Fig. 3).
Now it’s the time to go through those and check for the interesting ones. Once this is done, a list of relevant publications can be generated and downloaded using the interactive view of the last component in the workflow.
Note: This workflow uploaded to KNIME Business Hub creates a web based application that doesn’t expect any workflow building skills from the user.
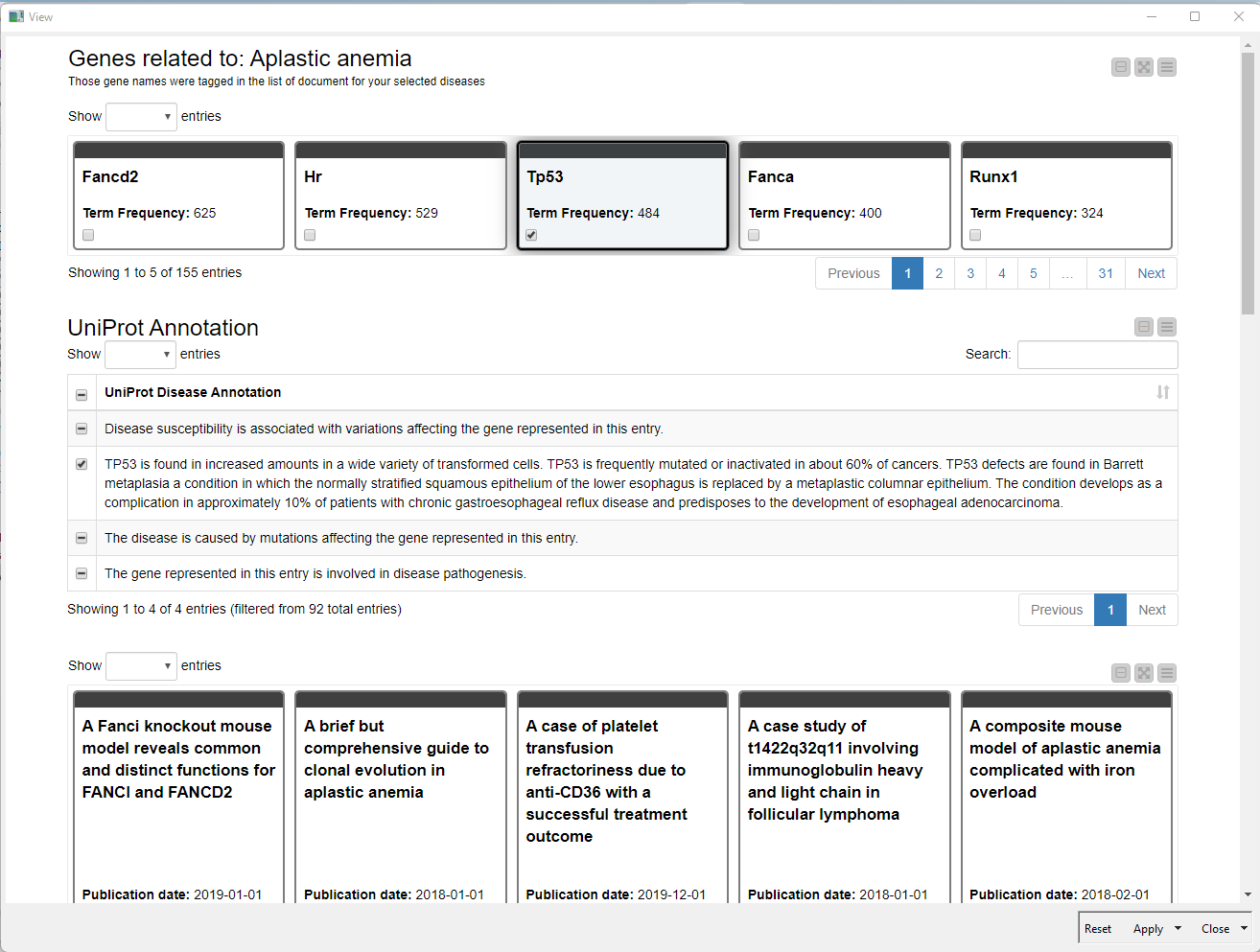
Example #2: Investigating Relationships of Specific Diseases & Genes
With this example we will show how easy it is to use dictionaries and ontologies and tag those terms on biomedical literature within KNIME Analytics Platform. It is in principle a similar example to the first one - we am to do a literate search - but let’s focus here on the reading part a bit more.
We start again with an input - this time a disease that needs to be entered or selected in the Autocomplete Widget Node. The node gets a list of diseases from the disease ontology that can be downloaded from the OBO foundry. To read the .rdf format we can use the Triple File Reader node.
The literature that will be downloaded with title and abstract will be in the next step tagged with genes to find out which genes are mentioned in publications with a certain disease. For this we connect with the UniPort SPARQL Endpoint using the SPARQL Endpoint node. With a dedicated query we get a list of genes and disease annotations from UniProt.
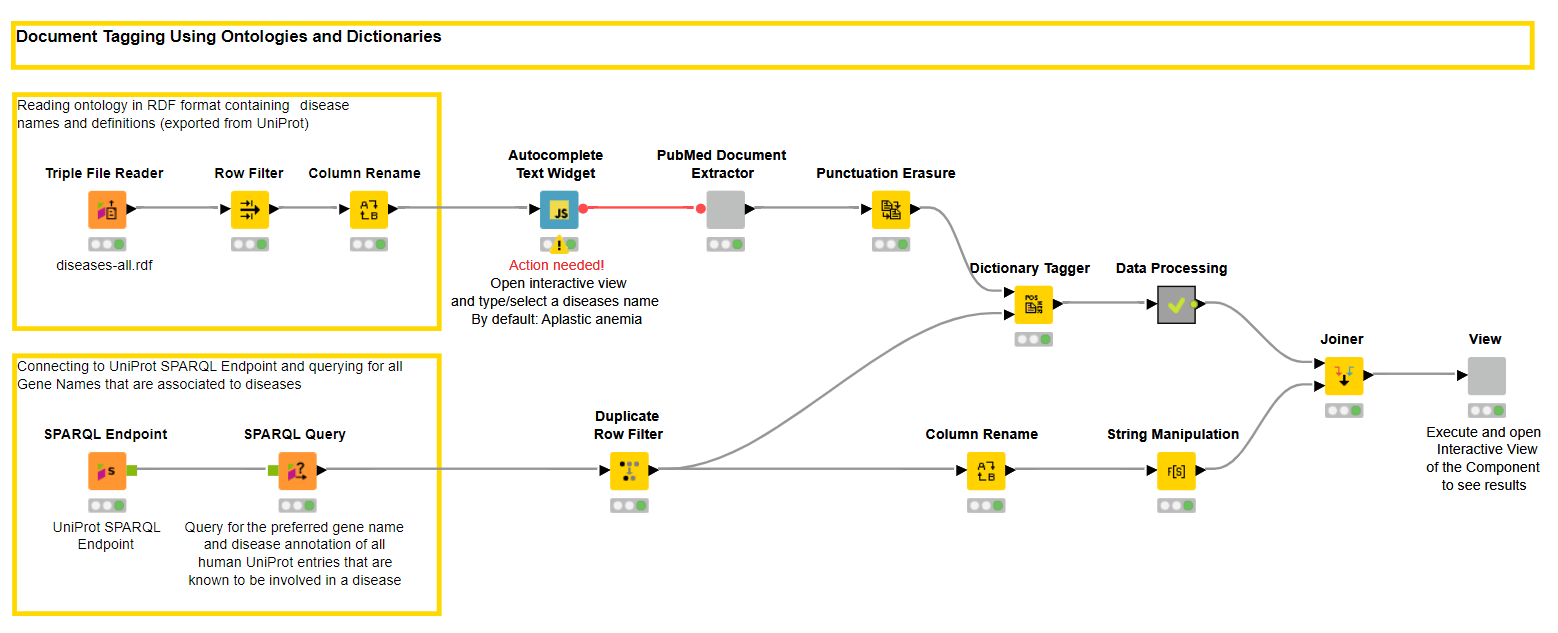
Once the terms and literature is loaded we map the genes against the publications using the Dictionary Tagger Node. With some more processing and data preparation we create a visualization within a component that allows us to interactively browse through the publication.
To make it easier to browse through the genes we counted the tags and those with the highest frequency are visible first. With this the user can decide if the known and already well researched, or the rare and maybe newly discovered genes are of interest for the disease under investigation.
Successfully Addressing Information Overload in Research Literature
In this blog post we discussed different approaches and options to undertake biomedical literature surveys using dictionaries and ontologies, tagging the abstracts according to those terms as well as displaying the results through an interactive visualization.
With this, we created a tool that can help to investigate for example hypotheses around adverse drug events with respect to ethnic backgrounds of a patient without going into different literature sources. Is this all that can be done using such approaches? Not at all! There are a lot more questions that could be tackled using such tools and extending the existing workflows with other sources, terms and ideas. This can ultimately lead to customizable and easily generalizable tools that can be applied to literature reviews in any domain.
